Research
Genetic architecture of speciation
The formation of new species underlies all of life’s biodiversity. Speciation is usually a gradual process, that can occur despite some interbreeding between the emerging gene pools. In fact our work on Heliconius butterflies has revealed that species can remain distinct despite ongoing, occasional hybridisation over a million years after they first started to diverge. So, what are the mechanisms that maintain species differences despite interbreeding?
One answer to this question is that natural selection is able to ‘weed out’ some foreign genes from each population, and thereby maintain some distinctness between the species.
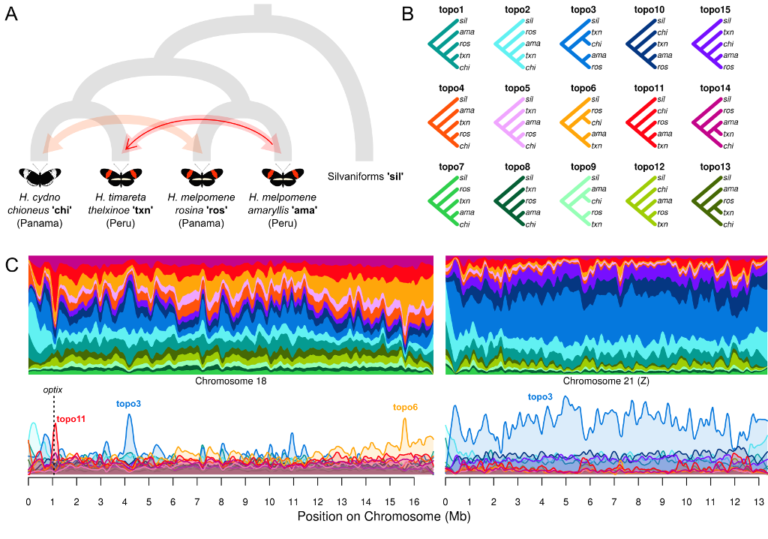
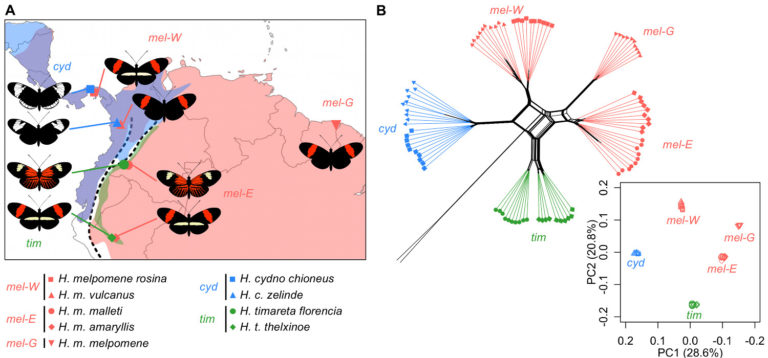
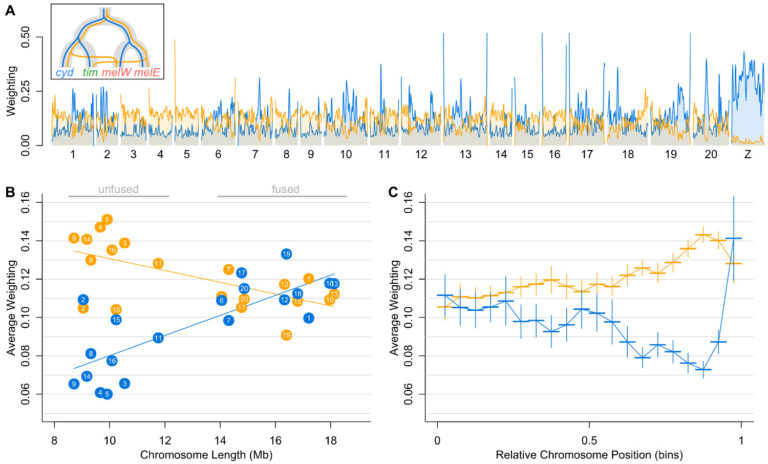
We have been asking where in the genome, and how, these species barriers form. One of the most striking patterns to emerge from our work, and studies in other taxa, is that the local recombination rate influences the effectiveness of species barriers. As a result, there are dramatic differences across the genome in how distinct two species are, and in some cases different parts of the genome even tell a different story of which species is most closely related to which.
Next, we want to know how repeatable these patterns are, and whether large scale changes to the organisation of the genome can change their role in speciation.
Genome structural evolution: causes and consequences
Genome structural changes – chromosomal inversions, translocations, fusions and fissions – have major evolutionary consequences. By altering recombination rates, they can contribute to species barriers as well as processes like genetic hitchhiking.
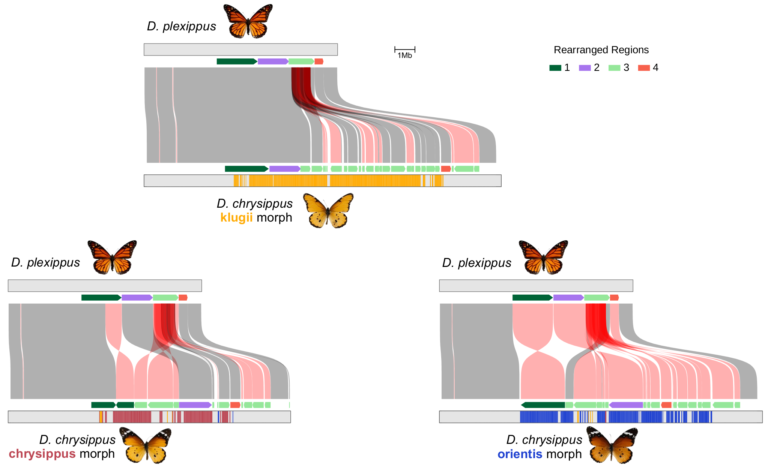
We have recently focused on a few case studies of major changes to genome architecture in butterflies and birds. In particular, in Danaus chrysippus, a single chromosome has undergone multiple rearrangements, giving rise to a colour patterning supergene, and more recently a neo-sex chromosome that has spread to several populations in East and Central Africa.
In ongoing work we are trying to better understand both the upstream causes and downstream consequences of structural evolution, both direct consequences on recombination rate and long term evolutionary consequences.
Computational tools for evolutionary genomics
With the increasing availability of genomic data for an ever growing range of taxa, our progress in understanding their evolution is now limited more by our ability to analyse and make sense of the data.
We are excited about developing computational tools for visualising and making inferences from genomic data.